The University of Michigan Next Generation Internet Implementation to Serve Visible Human Datasets
Alex Ade, Fred Bookstein, and Brian Athey
Introduction
The University of Michigan Visible Human (UMVH) project will use the Next Generation Internet (NGI) to serve Visible Human (VH) data to Health Science students, clinicians, basic science faculty, and researchers. Novel 2D and 3D navigational browsers will provide VH derived content in an educationally relevant manner (incorporating current trends in Advanced Distributed Learning technologies). Arbitrary slices, text, models, and flythroughs are being packaged into learning modules organized systemically or regionally to be delivered to many simultaneous users.
Collaboratory
The UMVH team is establishing a Visible Human Collaboratory for Medical Education and Training that connects researchers via the Next Generation Internet/Internet-2 backbone. The core groups include Stanford University, the University of Colorado Health Sciences, the Pittsburgh Supercomputing Center (PSC), the National Library of Medicine, and the Uniformed Service University of the Health Sciences. Working within the context of this larger collaboratory of VH researchers, UMVH promotes a set of open standards for datatypes and information related to the VH datasets. For example, such standards may include Extensible Markup Language (XML) for data interchange and Common Anatomical Markup Language (CAML) for data descriptors of anatomical terminology. Once adopted, other groups with NGI connectivity will be able to query the UMVH databases and access this information. Feedback from these groups will drive revisions and extensions.
Networking
The UMVH team is currently addressing two network issues. 1) Network testing between UM and PSC has uncovered bottlenecks within the framework and network architecture currently in place, and 2) the "last mile" problem, where data transmission to the client is typically throttled down at the final connection in the pathway. We predict that advances in high-speed network delivery such as DSL will eventually overcome this deficiency.
Past and current work
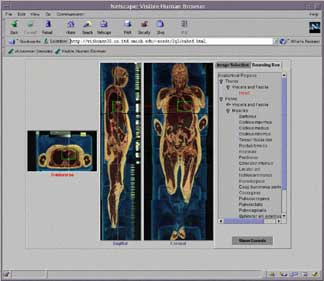
The UMVH project is building on two previous software development efforts. The first is a series of Web based browsers showing three orthogonal (transverse, sagittal, and coronal) views of the VH dataset in either high, medium, or low resolution. Enhancements to this software will include arbitrary slicing through the dataset, 3D model views showing model-plane and model-model relationships, network connectivity with the UM Anatomical database, and image-based/text-based queries (fig.1).

Fig.1
The second is the application, Edgewarp. Edgewarp is a stand-alone workstation package for manipulation of 2D and 3D biomedical images and related data structures by a combination of landmark location, thin-plate spline, and image unwarping and averaging. The new version of Edgewarp will eventually comprise three main modules: one for digitizing landmark point locations and curving data, one for displaying thin-plate splines in two and three dimensions, and one for image processing, unwarping, and averaging (fig 2).
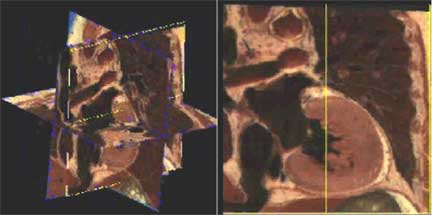
Fig. 2
Additionally, the UMVH team is working with Ethereal Technologies, Inc. to develop a real-time, immersive display system for the UMVH project. This will allow remote 3D visualizations of the VH dataset using standard PC graphics displayed onto a concave mirrored surface. Scenes are viewed in 3D without the use of conventional shutter glasses.
Conclusions
Software created during the UMVH project will address many issues, including scalability, quality of service (fault reduction and tolerance), and image understanding. The goal of this project is to architect an open software framework that accommodates future datasets and seamlessly integrates with data and software from other VH researchers within a larger collaboratory.